The inbreeding 2 experiment was planted in exPt1 in 2006 to determine how genetic drift is differentially affecting average fitness of remnant populations. In 2005, team members crossed common garden plants from seven remnant populations. There are three cross types: inbred (crossed to a half-sib; I), within population (randomly chosen; W), and between population (B). Each year, team members assess flowering phenology and fitness of Echinacea in the inb2 common garden.
In 2019, the team searched for Echinacea at 508 positions of the original 1443 positions planted in inb2. In total, we found 351 living plants. Four plants flowered in 2019 but only three produced achenes. Since 2006, 163 Echinacea in inb2 have flowered; they have produced a total of 336 flowering heads.
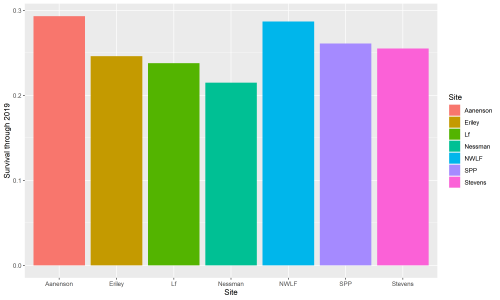
This winter, Riley Thoen is working on analyzing data and drafting a manuscript for inb2. In these endeavors, he found a small discrepancy in inb2 data: not all plants that were planted in the inb2 plot have a complete pedigree. Therefore, only a subset of the total can be used for analysis. A total of 1136 plants with a complete pedigree were planted in inb2, and of those, 277 were found alive in 2019. All four plants that flowered in 2019 have known pedigrees. A total of 138 plants of known pedigree have flowered and they have produced 284 total heads since the plot was planted in 2006. Surprisingly, within-remnant crosses have the lowest survival of all cross types, at 20%. Inbred crosses have 24% survival and between-remnant crosses have 30% survival. Riley is starting to push data analysis forwards and will certainly post updates on the flog when more discoveries are made!
For more summary plots, click these links:
Start Year: 2005 (crosses) and 2006 (planting)
Location: exPt1
Overlaps with: inb1, 1996 and 1997, common garden experiment, flowering phenology in experimental plots
Data/material collected: flowering phenology on the flowering plants (available in the exPt1 phenology data frames in the cgData repo), measure data (cgData repo), and harvested heads (data available in hh.2019 in the echinaceaLab package; heads in ACE protocol at CBG).
Products:
Shaw, R. G., S. Wagenius and C. J. Geyer. 2015. The susceptibility of Echinacea angustifolia to a specialist aphid: eco-evolutionary perspective on genotypic variation and demographic consequences. Journal of Ecology 103: 809-818. PDF
Kittelson, P., S. Wagenius, R. Nielsen, S. Qazi, M. Howe, G. Kiefer, and R. G. Shaw. 2015. Leaf functional traits, herbivory, and genetic diversity in Echinacea: Implications for fragmented populations. Ecology 96: 1877–1886. PDF

Leave a Reply